Assignment 5 (Oct 19)¶
Today we will learn to import data into Python, manipulate 2D tabular data (think spreadsheets) with pandas, and save our data to text files like CSV (comma-separated values).
Outline:
Intro to
pandasData I/O (reading and writing)
Selecting data
Manipulating data
Plotting
Readings (optional)¶
If you find this week’s material new or challenging, you may want to read through some or all the following resources while working on your assignment:
Below we have our regular imports, but this time with pandas which most people alias to pd. The Pandas package is included with Anaconda, so you shouldn’t need to install anything.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
Intro to Pandas¶
Pandas is a package at the heart of most data analysis tasks with Python. It is quick at opening, selecting, manipulating and plotting 2D data with columns and rows (like tables and spreadsheets) which pandas calls DataFrames.
Below we make a simple pandas.DataFrame to see what Pandas can do. One way to make a DataFrame is with two zipped lists, which we saw when making dictionaries in assignment 2.
fruit = ['apples', 'oranges', 'pears', 'bananas', 'kiwis', 'cherries']
count = [142, 119, 248, 84, 41, 187]
inventory = list(zip(fruit, count))
print(inventory)
[('apples', 142), ('oranges', 119), ('pears', 248), ('bananas', 84), ('kiwis', 41), ('cherries', 187)]
To turn this into a Pandas DataFrame, we call pd.DataFrame and specify the data as our zipped lists and also the columns names in order.
df = pd.DataFrame(data=inventory, columns=['Fruit', 'Count'])
print('df is type: ', type(df))
df
df is type: <class 'pandas.core.frame.DataFrame'>
| Fruit | Count | |
|---|---|---|
| 0 | apples | 142 |
| 1 | oranges | 119 |
| 2 | pears | 248 |
| 3 | bananas | 84 |
| 4 | kiwis | 41 |
| 5 | cherries | 187 |
Now we see we have a table version of our inventory data with named columns, “Fruit”, and “Count”. Let’s see what we can learn about these data.
df.head() # show first 5 rows
| Fruit | Count | |
|---|---|---|
| 0 | apples | 142 |
| 1 | oranges | 119 |
| 2 | pears | 248 |
| 3 | bananas | 84 |
| 4 | kiwis | 41 |
df.head(6) # show first n rows
| Fruit | Count | |
|---|---|---|
| 0 | apples | 142 |
| 1 | oranges | 119 |
| 2 | pears | 248 |
| 3 | bananas | 84 |
| 4 | kiwis | 41 |
| 5 | cherries | 187 |
len(df) # number of rows
6
print(df.columns) # column names
print(list(df.columns)) # as a normal list
Index(['Fruit', 'Count'], dtype='object')
['Fruit', 'Count']
df.dtypes # see data types for each column
Fruit object
Count int64
dtype: object
Each column in a DataFrame is assigned a type. We can change the type of a column with .astype. This is sometimes needed to ensure that you are working with numerical data, dates, etc.
df = df.astype({"Fruit": str, "Count": float})
df.dtypes
Fruit object
Count float64
dtype: object
A quirk of pandas is that it doesn’t recognize the str type. All we need to do is spell out "string" in this case.
df = df.astype({"Fruit": "string", "Count": float})
df.dtypes
Fruit string
Count float64
dtype: object
Data I/O: Importing a CSV¶
Data I/O (in/out) operations are an essential and often tedious part of analyzing scientific data. Pandas offers some help and can read simple structured data files with little trouble. These are mainly through the pd.read_ functions listed here, some of which include:
CSV:
pd.read_csv()Fixed-width:
pd.read_fwf()Excel:
pd.read_excel()and more…
Here we read in a CSV from a public URL. You can also read in CSVs on your local computer by passing in the path.
csv_url = "https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/05_Merge/Auto_MPG/cars1.csv"
df = pd.read_csv(csv_url)
df.head()
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | car | Unnamed: 9 | Unnamed: 10 | Unnamed: 11 | Unnamed: 12 | Unnamed: 13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307 | 130 | 3504 | 12.0 | 70 | 1 | chevrolet chevelle malibu | NaN | NaN | NaN | NaN | NaN |
| 1 | 15.0 | 8 | 350 | 165 | 3693 | 11.5 | 70 | 1 | buick skylark 320 | NaN | NaN | NaN | NaN | NaN |
| 2 | 18.0 | 8 | 318 | 150 | 3436 | 11.0 | 70 | 1 | plymouth satellite | NaN | NaN | NaN | NaN | NaN |
| 3 | 16.0 | 8 | 304 | 150 | 3433 | 12.0 | 70 | 1 | amc rebel sst | NaN | NaN | NaN | NaN | NaN |
| 4 | 17.0 | 8 | 302 | 140 | 3449 | 10.5 | 70 | 1 | ford torino | NaN | NaN | NaN | NaN | NaN |
It seems this CSV was imported with a bunch of empty columns. We can cut those off after importing, or adjust our read_csv to only include certain columns. This can be done by giving a list of indices to the usecols argument, but a range works just as well. Here we want the first 9 columns, meaning range(0, 9) in Python.
csv_url = "https://raw.githubusercontent.com/guipsamora/pandas_exercises/master/05_Merge/Auto_MPG/cars1.csv"
df = pd.read_csv(csv_url, usecols=range(0, 9))
df.head()
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | car | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307 | 130 | 3504 | 12.0 | 70 | 1 | chevrolet chevelle malibu |
| 1 | 15.0 | 8 | 350 | 165 | 3693 | 11.5 | 70 | 1 | buick skylark 320 |
| 2 | 18.0 | 8 | 318 | 150 | 3436 | 11.0 | 70 | 1 | plymouth satellite |
| 3 | 16.0 | 8 | 304 | 150 | 3433 | 12.0 | 70 | 1 | amc rebel sst |
| 4 | 17.0 | 8 | 302 | 140 | 3449 | 10.5 | 70 | 1 | ford torino |
The pandas read_csv can also read in other character delimited file formats, e.g. space or tab-separated (TSV) files. All we need to do is change the delimiter. Ex.
df = pd.read_csv(f, delimiter='\t') # Tab-separated values
df = pd.read_csv(f, delimiter='\s+') # Whitespace separated
[Assignment 1]¶
In this assignment, you will practice reading in several datasets of different formats.
1a) CSV (comma-separated)¶
We can read in CSV files like above with pd.read_csv. Try it on a5data.csv in the cell provided below (either go to the link and copy the url or right-click then copy link url to use in pd.read_csv).
1b) TSV (tab-separated)¶
To read in TSV files, we can also use pd.read_csv with the delimiter='\t' argument. Try it on a5data.tsv in the cell provided below.
1c) Excel (.xls, .xlsx)¶
To read in Excel files, we use pd.read_excel. Try it on a5data.xlsx in the cell provided below.
1d) ASCII text file¶
To read in text files with entries separated by some amount of whitespace (tabs, spaces, etc), we can use pd.read_csv with the delimiter='\s+' argument. Try it on a5data.txt in the cell provided below.
1e) ASCII text file with header rows¶
To have pandas skip rows while importing, we can use the skiprows argument of pd.read_csv. Try it on a5data_headers.txt in the space provided below.
1f) BONUS: FITS table file¶
(Optional): For this question, you will need to read in tabular data in a .fits file with astropy.table.Table() (note: this only works for FITS tables, not images). Once you’ve read in the table with astropy, convert it to a pd.DataFrame with the .to_pandas() method. You will need the following line to import astropy Table:
from astropy.table import Table.
Try it on a5data.fits in the cell provided below. See other file formats supported by astropy here.
This data is from a paper by Shih-Yun Tang et al. (2019)
# [1a code below]
print(df['Gmag'].max())
# [1b code below]
print(df['Gmag'].max())
# [1c code below]
print(df['Gmag'].max())
# [1d code below]
print(df['Gmag'].max())
# [1e code below]
print(df['Gmag'].max())
# [1f code below]
from astropy.table import Table
print(df['Gmag'].max())
Selecting data with Pandas¶
Pandas offers several ways to access and filter data in DataFrames. The most basic are [], .loc[] and .iloc[].
Selecting with square brackets [] and dot .¶
Selecting with square brackets [] gives us the column that we specify.
df['car'] # Select a column by its name
0 chevrolet chevelle malibu
1 buick skylark 320
2 plymouth satellite
3 amc rebel sst
4 ford torino
...
193 ford maverick
194 amc hornet
195 chevrolet chevette
196 chevrolet woody
197 vw rabbit
Name: car, Length: 198, dtype: object
Alternatively, we can access DataFrame columns with .columnname.
df.car
0 chevrolet chevelle malibu
1 buick skylark 320
2 plymouth satellite
3 amc rebel sst
4 ford torino
...
193 ford maverick
194 amc hornet
195 chevrolet chevette
196 chevrolet woody
197 vw rabbit
Name: car, Length: 198, dtype: object
There is no major difference between the two methods, it’s a matter of personal preference. The dot (.) version is less typing, but is harder to use when you have your column name stored as a variable, for example:
col = 'mpg'
df[col] # This works!
# df.col # This doesn't! AttributeError since "col" is not a column name
0 18.0
1 15.0
2 18.0
3 16.0
4 17.0
...
193 24.0
194 22.5
195 29.0
196 24.5
197 29.0
Name: mpg, Length: 198, dtype: float64
Selecting with .loc[]¶
We can also select labelled columns or rows in our DataFrames with .loc[].
df.loc['car']
# KeyError
While DataFrame .loc[] allows us to select by the row name and column name, we have to do it in the proper order [rows, columns]. To select the car columns, we can use the : as a placeholder meaning “all rows”.
df.loc[:, 'car']
0 chevrolet chevelle malibu
1 buick skylark 320
2 plymouth satellite
3 amc rebel sst
4 ford torino
...
193 ford maverick
194 amc hornet
195 chevrolet chevette
196 chevrolet woody
197 vw rabbit
Name: car, Length: 198, dtype: object
So now we know df['car'] is equivalent to df.loc[:, 'car']. We can also select named rows of data by their Index. Right now our rows are just numbered, but a DataFrame Index could have any unique label (usually int or str).
df.loc[0] # First row of data
mpg 18.0
cylinders 8
displacement 307
horsepower 130
weight 3504
acceleration 12.0
model 70
origin 1
car chevrolet chevelle malibu
Name: 0, dtype: object
We can also select a single cell of data by specifying the row and column.
df.loc[11, 'car'] # The car in row 11
"plymouth 'cuda 340"
df.loc[29, 'weight'] # The weight in row 29
2130
We can also select multiple Index or columns by passing a list to .loc[].
df.loc[1, ['car', 'horsepower', 'cylinders']]
car buick skylark 320
horsepower 165
cylinders 8
Name: 1, dtype: object
df.loc[range(12, 17), 'car']
12 chevrolet monte carlo
13 buick estate wagon (sw)
14 toyota corona mark ii
15 plymouth duster
16 amc hornet
Name: car, dtype: object
If we select along one row or column, we get a 1D slice of our DataFrame. We can also select multiple indices and columns to get a smaller DataFrame. The result keeps the same order as the rows and columns we specify.
df.loc[[3, 2, 7, 1], ['car', 'cylinders', 'horsepower']]
| car | cylinders | horsepower | |
|---|---|---|---|
| 3 | amc rebel sst | 8 | 150 |
| 2 | plymouth satellite | 8 | 150 |
| 7 | plymouth fury iii | 8 | 215 |
| 1 | buick skylark 320 | 8 | 165 |
A common task is to make a subset of our DataFrame with only the columns we need. We can do this easily with .loc[].
sub_df = df.loc[:, ['car', 'cylinders', 'horsepower', 'weight', 'mpg', 'acceleration']]
sub_df.head()
| car | cylinders | horsepower | weight | mpg | acceleration | |
|---|---|---|---|---|---|---|
| 0 | chevrolet chevelle malibu | 8 | 130 | 3504 | 18.0 | 12.0 |
| 1 | buick skylark 320 | 8 | 165 | 3693 | 15.0 | 11.5 |
| 2 | plymouth satellite | 8 | 150 | 3436 | 18.0 | 11.0 |
| 3 | amc rebel sst | 8 | 150 | 3433 | 16.0 | 12.0 |
| 4 | ford torino | 8 | 140 | 3449 | 17.0 | 10.5 |
One limitation of .loc[] is that we can’t select a range of str, in this case our columns. We can choose a range of columns with .iloc[] instead.
sub_df = df.loc[:, range(4)]
# KeyError
Selecting with .iloc[]¶
With .iloc[], we can select data by the numbered Index or columns.
sub_df = df.iloc[:, range(4)]
sub_df.head()
| mpg | cylinders | displacement | horsepower | |
|---|---|---|---|---|
| 0 | 18.0 | 8 | 307 | 130 |
| 1 | 15.0 | 8 | 350 | 165 |
| 2 | 18.0 | 8 | 318 | 150 |
| 3 | 16.0 | 8 | 304 | 150 |
| 4 | 17.0 | 8 | 302 | 140 |
So in pandas, both the order of the rows and columns and their labels are important! We can select by labels using .loc and by integer index order using .iloc.
df2 = df.loc[[3, 2, 7, 1], ['car', 'cylinders', 'horsepower']]
df2.head()
| car | cylinders | horsepower | |
|---|---|---|---|
| 3 | amc rebel sst | 8 | 150 |
| 2 | plymouth satellite | 8 | 150 |
| 7 | plymouth fury iii | 8 | 215 |
| 1 | buick skylark 320 | 8 | 165 |
sub_df.iloc[:, 0] # mpg was 1st column
0 18.0
1 15.0
2 18.0
3 16.0
4 17.0
...
193 24.0
194 22.5
195 29.0
196 24.5
197 29.0
Name: mpg, Length: 198, dtype: float64
df2.iloc[:, 0] # car is 1st column
3 amc rebel sst
2 plymouth satellite
7 plymouth fury iii
1 buick skylark 320
Name: car, dtype: object
Setting a new index in pandas¶
We can choose a column of unique values to be our new pandas index. Here, let’s use the car names.
df2 = df.set_index('car')
df2.head()
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | |
|---|---|---|---|---|---|---|---|---|
| car | ||||||||
| chevrolet chevelle malibu | 18.0 | 8 | 307 | 130 | 3504 | 12.0 | 70 | 1 |
| buick skylark 320 | 15.0 | 8 | 350 | 165 | 3693 | 11.5 | 70 | 1 |
| plymouth satellite | 18.0 | 8 | 318 | 150 | 3436 | 11.0 | 70 | 1 |
| amc rebel sst | 16.0 | 8 | 304 | 150 | 3433 | 12.0 | 70 | 1 |
| ford torino | 17.0 | 8 | 302 | 140 | 3449 | 10.5 | 70 | 1 |
Now we can select certain rows by the car name using .loc.
df2.loc['amc rebel sst']
mpg 16.0
cylinders 8
displacement 304
horsepower 150
weight 3433
acceleration 12.0
model 70
origin 1
Name: amc rebel sst, dtype: object
Be careful when working with a numerical Index(). You can run into confusing situations where the index associated with your rows is not in the same order as your rows. Recall the earlier example
df3 = df.loc[[3, 2, 7, 1], ['car', 'cylinders', 'horsepower']]
df3.head()
| car | cylinders | horsepower | |
|---|---|---|---|
| 3 | amc rebel sst | 8 | 150 |
| 2 | plymouth satellite | 8 | 150 |
| 7 | plymouth fury iii | 8 | 215 |
| 1 | buick skylark 320 | 8 | 165 |
Here, our Index has the numbers [3, 2, 7, 1], in that order as we can see with the .index.
df3.index
Int64Index([3, 2, 7, 1], dtype='int64')
This means we can select any values in our index with .loc, or any row number from 0 to 3 with .iloc.
print(df3.loc[7]) # Index 7
print(df3.iloc[2]) # Row 3
car plymouth fury iii
cylinders 8
horsepower 215
Name: 7, dtype: object
car plymouth fury iii
cylinders 8
horsepower 215
Name: 7, dtype: object
But here we need to be careful because we can pick the 1st row with .iloc[0], but there is no row with index 0 (remember our only indices are [3, 2, 7, 1]).
print(df3.iloc[0])
# print(df3.loc[0]) # KeyError
car amc rebel sst
cylinders 8
horsepower 150
Name: 3, dtype: object
If the numerical Index assigned by pandas doesn’t matter and we don’t want this situation where there are gaps or different orders of our rows, we can call .reset_index(drop=True). This will renumber our DataFrame rows starting from 0.
df3.head()
| car | cylinders | horsepower | |
|---|---|---|---|
| 3 | amc rebel sst | 8 | 150 |
| 2 | plymouth satellite | 8 | 150 |
| 7 | plymouth fury iii | 8 | 215 |
| 1 | buick skylark 320 | 8 | 165 |
df4 = df3.reset_index(drop=True)
df4.head()
| car | cylinders | horsepower | |
|---|---|---|---|
| 0 | amc rebel sst | 8 | 150 |
| 1 | plymouth satellite | 8 | 150 |
| 2 | plymouth fury iii | 8 | 215 |
| 3 | buick skylark 320 | 8 | 165 |
Summary: Selecting from DataFrames¶
Keep in mind:
['colname']or.colnameselects columns.loc[]selects by a label (i.e., a value indf.indexordf.columns).iloc[]selects by a numbered index in row / column order, starting at 0
Adding a new column¶
We can add columns to a DataFrame with DataFrame['col'] = values. We need to make sure the new values are the same number of rows as the DataFrame. An easy way to do this is to derive columns from existing DataFrame columns.
# Convert miles per gallons to liters per 100 km
df['lp100km'] = round(235.214583 / df['mpg'], 2)
df.head()
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | car | lp100km | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307 | 130 | 3504 | 12.0 | 70 | 1 | chevrolet chevelle malibu | 13.07 |
| 1 | 15.0 | 8 | 350 | 165 | 3693 | 11.5 | 70 | 1 | buick skylark 320 | 15.68 |
| 2 | 18.0 | 8 | 318 | 150 | 3436 | 11.0 | 70 | 1 | plymouth satellite | 13.07 |
| 3 | 16.0 | 8 | 304 | 150 | 3433 | 12.0 | 70 | 1 | amc rebel sst | 14.70 |
| 4 | 17.0 | 8 | 302 | 140 | 3449 | 10.5 | 70 | 1 | ford torino | 13.84 |
Saving to a file¶
It’s nice to manipulate and plot our data in Python, but we often want to save the results of our efforts for later.
The simplest way to do so is with pandas .to_csv. Pandas saves the Index by default, but since we just have default indices, you can just omit the index from the csv with index=False.
fname = '/full/path/to/cars.csv' # <- change to real path
# df.to_csv(fname, index=False) # <- uncomment to save to file
Other useful DataFrame methods¶
df.tail() # last 5 rows
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | car | lp100km | |
|---|---|---|---|---|---|---|---|---|---|---|
| 193 | 24.0 | 6 | 200 | 81 | 3012 | 17.6 | 76 | 1 | ford maverick | 9.80 |
| 194 | 22.5 | 6 | 232 | 90 | 3085 | 17.6 | 76 | 1 | amc hornet | 10.45 |
| 195 | 29.0 | 4 | 85 | 52 | 2035 | 22.2 | 76 | 1 | chevrolet chevette | 8.11 |
| 196 | 24.5 | 4 | 98 | 60 | 2164 | 22.1 | 76 | 1 | chevrolet woody | 9.60 |
| 197 | 29.0 | 4 | 90 | 70 | 1937 | 14.2 | 76 | 2 | vw rabbit | 8.11 |
df.describe() # summary statistics on each numerical column
| mpg | cylinders | displacement | weight | acceleration | model | origin | lp100km | |
|---|---|---|---|---|---|---|---|---|
| count | 198.000000 | 198.000000 | 198.000000 | 198.000000 | 198.000000 | 198.000000 | 198.000000 | 198.000000 |
| mean | 19.719697 | 5.898990 | 223.469697 | 3177.888889 | 15.005556 | 72.818182 | 1.439394 | 13.029293 |
| std | 5.814254 | 1.785417 | 115.181017 | 934.783733 | 2.872382 | 1.865332 | 0.708085 | 3.924258 |
| min | 9.000000 | 3.000000 | 68.000000 | 1613.000000 | 8.000000 | 70.000000 | 1.000000 | 6.720000 |
| 25% | 15.000000 | 4.000000 | 113.250000 | 2302.500000 | 13.000000 | 71.000000 | 1.000000 | 9.650000 |
| 50% | 19.000000 | 6.000000 | 228.000000 | 3030.000000 | 15.000000 | 73.000000 | 1.000000 | 12.380000 |
| 75% | 24.375000 | 8.000000 | 318.000000 | 4080.750000 | 16.800000 | 74.000000 | 2.000000 | 15.680000 |
| max | 35.000000 | 8.000000 | 455.000000 | 5140.000000 | 23.500000 | 76.000000 | 3.000000 | 26.130000 |
fruit = ['apples', 'apples', 'pears', 'bananas', 'kiwis', 'cherries']
count = [142, 142, 248, 84, None, 187]
fruit_df = pd.DataFrame(zip(fruit, count), columns=['Fruit', 'Count'])
fruit_df.head()
| Fruit | Count | |
|---|---|---|
| 0 | apples | 142.0 |
| 1 | apples | 142.0 |
| 2 | pears | 248.0 |
| 3 | bananas | 84.0 |
| 4 | kiwis | NaN |
fruit_df = fruit_df.fillna(0) # fill missing values with 0
fruit_df.head()
| Fruit | Count | |
|---|---|---|
| 0 | apples | 142.0 |
| 1 | apples | 142.0 |
| 2 | pears | 248.0 |
| 3 | bananas | 84.0 |
| 4 | kiwis | 0.0 |
fruit_df = fruit_df.drop_duplicates()
fruit_df.head()
| Fruit | Count | |
|---|---|---|
| 0 | apples | 142.0 |
| 2 | pears | 248.0 |
| 3 | bananas | 84.0 |
| 4 | kiwis | 0.0 |
| 5 | cherries | 187.0 |
Filtering DataFrames¶
We often want to select data within a certain range or on a specific condition. There are a couple ways to do this. The simplest (but slowest and most error-prone) way is to use a loop.
Say we want to select only cars with 4 cylinders. Here we use df.iterrows() which will return the Index of the row and the row itself on each iteration in the loop.
cars_4cyl = []
for i, row in df.iterrows():
if row['cylinders'] == 4:
cars_4cyl.append(row['car'])
print(cars_4cyl[:5])
['toyota corona mark ii', 'datsun pl510', 'volkswagen 1131 deluxe sedan', 'peugeot 504', 'audi 100 ls']
Now we have a list of 4 cylinder cars, but we’ve lost all other information stored about them in the DataFrame. We would need to make a separate list for each column we want to store.
Instead of using a loop, we can do the vectorized solution which keeps our data in a DataFrame and selects info based on our condition (row['cylinders'] == 4).
df_4cyl = df[df.cylinders == 4]
# df_4cyl = df[df['cylinders'] == 4] # Equivalent to the above
print('Max cylinders in DataFrame', df_4cyl.cylinders.max())
df_4cyl.head()
Max cylinders in DataFrame 4
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | car | lp100km | |
|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 24.0 | 4 | 113 | 95 | 2372 | 15.0 | 70 | 3 | toyota corona mark ii | 9.80 |
| 18 | 27.0 | 4 | 97 | 88 | 2130 | 14.5 | 70 | 3 | datsun pl510 | 8.71 |
| 19 | 26.0 | 4 | 97 | 46 | 1835 | 20.5 | 70 | 2 | volkswagen 1131 deluxe sedan | 9.05 |
| 20 | 25.0 | 4 | 110 | 87 | 2672 | 17.5 | 70 | 2 | peugeot 504 | 9.41 |
| 21 | 24.0 | 4 | 107 | 90 | 2430 | 14.5 | 70 | 2 | audi 100 ls | 9.80 |
To break down what we are doing here, we can look at what df.cylinders == 4 gives us.
df.cylinders == 4
0 False
1 False
2 False
3 False
4 False
...
193 False
194 False
195 True
196 True
197 True
Name: cylinders, Length: 198, dtype: bool
This is a boolean index, meaning it will be True or False based on the condition we supplied (whether there are 4 cylinders or not).
When we pass a boolean index to a DataFrame in square brackets ([]), it’s similar to supplying specific labels or indices as we learned above. The only difference is it will select only the rows in the DataFrame that have True, e.g. rows where the car has 4 cylinders.
We can supply conditions with any of the comparison operators (==, <, <=, >, >=). Say we want to find cars that get the fewest miles per gallon (mpg < 12):
df[df.mpg < 12]
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | car | lp100km | |
|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 10.0 | 8 | 360 | 215 | 4615 | 14.0 | 70 | 1 | ford f250 | 23.52 |
| 26 | 10.0 | 8 | 307 | 200 | 4376 | 15.0 | 70 | 1 | chevy c20 | 23.52 |
| 27 | 11.0 | 8 | 318 | 210 | 4382 | 13.5 | 70 | 1 | dodge d200 | 21.38 |
| 28 | 9.0 | 8 | 304 | 193 | 4732 | 18.5 | 70 | 1 | hi 1200d | 26.13 |
| 67 | 11.0 | 8 | 429 | 208 | 4633 | 11.0 | 72 | 1 | mercury marquis | 21.38 |
| 103 | 11.0 | 8 | 400 | 150 | 4997 | 14.0 | 73 | 1 | chevrolet impala | 21.38 |
| 124 | 11.0 | 8 | 350 | 180 | 3664 | 11.0 | 73 | 1 | oldsmobile omega | 21.38 |
We can also string conditions together by separating them with parentheses (()) and the union & or intersection | operators.
Union (
&): select where condition1 AND condition2 areTrueIntersection (
|): select where condition1 OR condition2 areTrue
light_high_accel = df[(df.weight < 2000) & (df.acceleration > 20)]
light_high_accel.head()
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | car | lp100km | |
|---|---|---|---|---|---|---|---|---|---|---|
| 19 | 26.0 | 4 | 97 | 46 | 1835 | 20.5 | 70 | 2 | volkswagen 1131 deluxe sedan | 9.05 |
| 56 | 26.0 | 4 | 91 | 70 | 1955 | 20.5 | 71 | 1 | plymouth cricket | 9.05 |
| 102 | 26.0 | 4 | 97 | 46 | 1950 | 21.0 | 73 | 2 | volkswagen super beetle | 9.05 |
| 131 | 32.0 | 4 | 71 | 65 | 1836 | 21.0 | 74 | 3 | toyota corolla 1200 | 7.35 |
heavy_or_8cyl = df[(df.weight > 3000) | (df.cylinders == 8)]
heavy_or_8cyl.tail()
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | car | lp100km | |
|---|---|---|---|---|---|---|---|---|---|---|
| 190 | 14.5 | 8 | 351 | 152 | 4215 | 12.8 | 76 | 1 | ford gran torino | 16.22 |
| 191 | 22.0 | 6 | 225 | 100 | 3233 | 15.4 | 76 | 1 | plymouth valiant | 10.69 |
| 192 | 22.0 | 6 | 250 | 105 | 3353 | 14.5 | 76 | 1 | chevrolet nova | 10.69 |
| 193 | 24.0 | 6 | 200 | 81 | 3012 | 17.6 | 76 | 1 | ford maverick | 9.80 |
| 194 | 22.5 | 6 | 232 | 90 | 3085 | 17.6 | 76 | 1 | amc hornet | 10.45 |
Finally, we can take the opposite of a comparison with a preceding negation operator (~).
# Use str.contains to match a sub-string
ford_cars = df[df.car.str.contains('ford')]
ford_cars.head()
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | car | lp100km | |
|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 17.0 | 8 | 302 | 140 | 3449 | 10.5 | 70 | 1 | ford torino | 13.84 |
| 5 | 15.0 | 8 | 429 | 198 | 4341 | 10.0 | 70 | 1 | ford galaxie 500 | 15.68 |
| 17 | 21.0 | 6 | 200 | 85 | 2587 | 16.0 | 70 | 1 | ford maverick | 11.20 |
| 25 | 10.0 | 8 | 360 | 215 | 4615 | 14.0 | 70 | 1 | ford f250 | 23.52 |
| 32 | 25.0 | 4 | 98 | ? | 2046 | 19.0 | 71 | 1 | ford pinto | 9.41 |
high_accel_not_ford = df[(df.acceleration > 15) & ~df.car.str.contains('ford')]
high_accel_not_ford.head()
| mpg | cylinders | displacement | horsepower | weight | acceleration | model | origin | car | lp100km | |
|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 22.0 | 6 | 198 | 95 | 2833 | 15.5 | 70 | 1 | plymouth duster | 10.69 |
| 16 | 18.0 | 6 | 199 | 97 | 2774 | 15.5 | 70 | 1 | amc hornet | 13.07 |
| 19 | 26.0 | 4 | 97 | 46 | 1835 | 20.5 | 70 | 2 | volkswagen 1131 deluxe sedan | 9.05 |
| 20 | 25.0 | 4 | 110 | 87 | 2672 | 17.5 | 70 | 2 | peugeot 504 | 9.41 |
| 22 | 25.0 | 4 | 104 | 95 | 2375 | 17.5 | 70 | 2 | saab 99e | 9.41 |
Plotting with DataFrame.plot¶
Pandas gives you a convenient interface into plotting with the .plot method. It even labels your axes for you!
ax = df.plot(x='mpg', y='weight', kind='scatter')
plt.show()
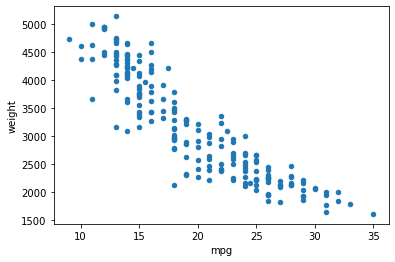
Of course, we can customize our subplot with the ax object it returned, as usual.
ax = df.mpg.plot(kind='hist')
ax.set_title('Fuel economy (mpg)')
plt.show()

[Assignment 2]¶
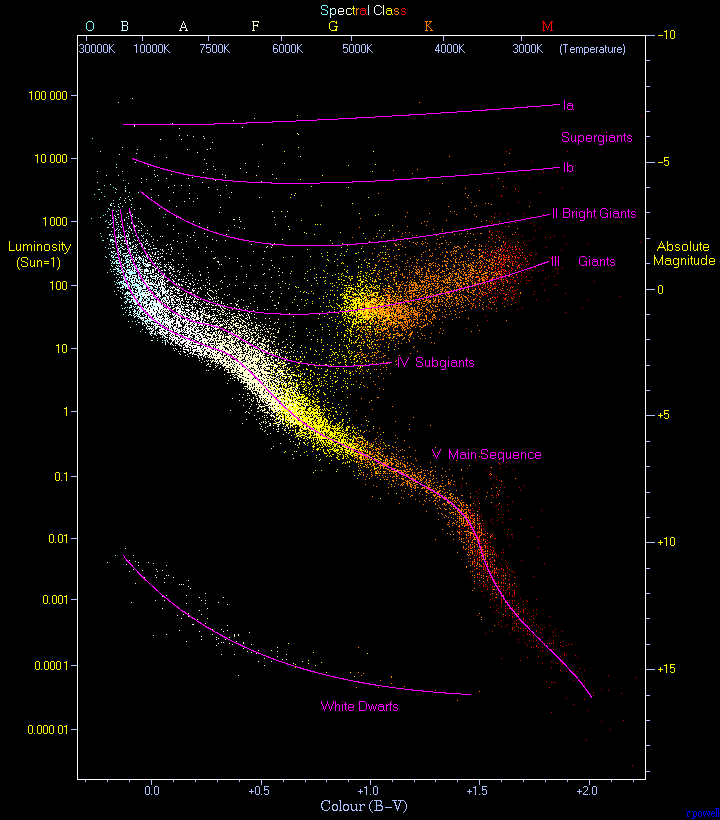
In this assignment you will plot a Hertzprung-Russell (H-R) diagram which shows stellar magnitude over color and can tell us about the populations of stars we are observing (see image below).
In the data we imported in assignment 1 (choose any import link / method you’d like), you will find DataFrame columns for G-band magnitude (Gmag), and color as magnitude of the blue part (BPmag), and magnitude of the red part (RPmag).
On H-R diagrams (see image), we want absolute magnitude on the y-axis with the brightest stars at the top (note: low stellar magnitudes are brighter). We also want to plot color as BPmag - RPmag on the x-axis.
In our DataFrame, we only have an apparent magnitude, so we will need to derive the absolute magnitude before we can make our plot.
We can compute Absolute magnitude (\(M\)) if we know the apparent magnitude (\(m\)) and the distance modulus (\(\mu\)) by:
The distance modulus can be calculated from the distance to a star in parsecs (pc):
Finally, we can determine a star’s distance in parsecs from the measured parallax in milliarcseconds (mas) by:
Note parallax is Plx in our DataFrame. Now we know all we need to make an H-R diagram and identify some stars!
2a) Convert apparent magnitude to absolute magnitude¶
Using the equations in the question, derive the absolute G magnitude (\(M\)) from the apparent G magnitude for each star in the DataFrame. Hint: you will need columns Gmag, BPmag, RPmag, and Plx.
In each step, save the result as a new column in your DataFrame, for example:
df['distance_modulus'] = # calculation for distance modulus here
Your final DataFrame should have columns for distance_modulus, distance_parsecs, and absolute_magnitude.
2b) Calculate the color¶
With the data provided, we can find color using blue part (BPmag) - red part (RPmag). Save this color difference as a column of your DataFrame.
2c) Plot the HR diagram¶
Using pandas DataFrame.plot(x='color', y='absolute_magnitude'), plot the H-R diagram. Make sure to label your axes!
2d) Identify and select the white dwarfs¶
White dwarfs on the H-R diagram are outliers in the lower left.
Using a conditional statement (or two) on absolute magnitude and color, select all white dwarfs and save them to their own DataFrame. Call .describe() on this new DataFrame to get summary statistics of the white dwarfs.
2e) Identify and select the giants¶
Giants on the H-R diagram are outliers in the top middle to right section of the diagram.
Using a conditional statement (or two) on absolute magnitude and color, select all giants and save them to their own DataFrame. Call .describe() on this new DataFrame to get summary statistics of the giants.
2f) Plot the white dwarfs and giants separately¶
Using your white dwarf and supergiant DataFrames, plot each on the same axes as your H-R diagram with distinct markers and colors (hint: try changing the markerstyle, color, markersize options of .plot()).
To plot all on the same axes, make a subplot and pass in ax to each .plot call, e.g.
fig, ax = plt.subplots()
df.plot(ax=ax)
df2.plot(ax=ax)
...
2g) Write the white dwarfs and giants to CSVs¶
Write your white dwarf and giants DataFrames to CSV files with .to_csv(), using the index=False option. Include these CSV files in the assignment 5 directory and in your commit / PR.
2h) BONUS: Label the white dwarfs and giants¶
Research the ax.annotate() method of matplotlib axes and label the white dwarf and supergiant points on your plot as “white dwarf” and “supergiant” with arrows pointing to each. As always, bonus points for any other plot aesthetics you add!
# [your code here]
